International Journal of Pharmaceutical Sciences and Developmental Research
Oropharyngeal Pathogenic Bacteria: Carriage, Antimicrobial Susceptibility Pattern and Associated Risk Factors among Febrile Patients
Merete Tesfaye Wondimu1, Teklu Shiferaw Simbo2, Tolossa Eticha Chaka3, Haile Abera Lemi4*, Biruk Yeshitila5, Mekonnen Teferi6, Kemal Jemal7 and Worku Bedada8*
2Assistant Professor of Medical Microbiology, Department of Biomedical Sciences, Adama Hospital Medical College, Adama, Oromia, Ethiopia
3Associate Professor of Pediatrics, Department of Pediatrics, Adama Hospital Medical College, Adama, Oromia, Ethiopia
4Medical Laboratory Technologist, Armauer Hansen Research Institute, Addis Ababa, Ethiopia
5PhD Candidate and Researcher, Armauer Hansen Research Institute, Addis Ababa, Ethiopia
6Senior Researcher, Armauer Hansen Research Institute, Addis Ababa Ethiopia
7Assistant Professor of Nursing, Department of Nursing, College of Health Sciences, Salale University, Oromia, Ethiopia
8Assistant Professor of Pharmacology, Department of Biomedical Sciences, Adama Hospital Medical College, Adama, Oromia, Ethiopia
Haile Abera Lemi, Armauer Hansen Research Institute, Addis Ababa, Ethiopia, Email: [email protected]
Cite this as
Wondimu MT, Simbo TS, Chaka TE, Lemi HA, Bedada W, et al. (2022) Oropharyngeal Pathogenic Bacteria: Carriage, Antimicrobial Susceptibility Pattern and Associated Risk Factors among Febrile Patients. Int J Pharm Sci Dev Res 8(1): 006-012 DOI: 10.17352/ijpsdr.000037Copyright License
© 2022 Wondimu MT, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: Pharyngeal and respiratory infections due to bacteria are global concerns especially because of the emergence of multi-drug resistant strains. The Oropharynx is one of the regions of the human body that is heavily colonized by microbial flora. So, the Oropharyngeal carriage is a major risk factor for an invasion and developing the disease. Therefore, this study was aimed at determining the carriage rate of potential pathogenic Oropharyngeal bacteria, associated risk factors, and antimicrobial susceptibility patterns among febrile patients at AHMC.
Methods: A Cross-sectional study was conducted from November 2018 to April 2019. A systematic random technique was implemented to collect Oropharyngeal samples from 253 selected febrile patients. Specimen collection and processing were done following standard bacteriological procedures in Adama Public Health, Research, and Referral Laboratory. Antimicrobial susceptibility testing was done using the disk diffusion method in accordance with the Clinical and Laboratory Standard Institutes guideline. Data were analyzed using SPSS version 20.
Results: Overall oropharyngeal bacteria carriage rate was 75.1%. A total of 190 bacterial organisms were isolated with 96 Coagulase-Negative Staphylococci (CNS), 55 Streptococcus pyogenes, and 19 Staphylococcus aureus were dominant isolates among gram-positive. Klebsiella pneumoniae, Neisseria meningitides, Moraxella catarrhalis, and Escherichia coli were among gram-negative bacteria isolates. Living in urban, cigarette smoking, and poor oral hygiene was significantly associated with the Oropharyngeal bacterial carriage rate. The majority of CNS, S.aureus, and S. pyogenes were resistant to different classes of antibiotics.
Conclusion: Potentially pathogenic Oropharyngeal bacterial colonization rate was high. Place of living and behavioral factors are risk factors that were associated with bacterial colonization of the oropharynx. Several bacterial isolates were resistant to frequently prescribed antibiotic classes. Therefore, determining the antimicrobial susceptibility pattern of the isolates is important to intervene and prevent the risk of infection and also the emergence of multiple drug-resistant strains and disease management.
Introduction
The human Oropharynx is an aerated, mucus-rich chamber, comprising the posterior pharyngeal wall and bilateral structures of the palatine tonsils. The contiguity of the oropharynx exposes to the microbes that originated from the saliva and the external environments [1]. The upper respiratory tract is an ecological niche for many microbial species, including Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Neisseria meningitides, Haemophilus influenza, and Moraxella catarrhalis [2]. These pathogenic bacteria can colonize the Oropharyngeal epithelium of apparently healthy individuals.
The microbial flora of the human oral cavity is highly diverse, consisting of main bacteria (gram-positive cocci and aerobic gram negatives), fungi, and protozoa. These microbes play an important role in preventing the colonization of other pathogenic microbes [3]. The disruption of the normal flora could trigger or influence the course of oral diseases and it could also be a source for systemic diseases. The oropharynx is a complex and dynamic environment; where pneumococcal interact with the host immune system and with other colonizing bacteria. In some cases, these interactions are mutually beneficial, whereas, in others, there is evidence for competition [2,4].
The World Health Organization estimates that respiratory infections account for 1.9 to 2.2 million childhood deaths annually, with 70% occurring in Africa and Southeast Asia [5]. Community Oropharynx carriage rates vary from 22% to 28% across the world [6]. S. pneumoniae, H. influenzae, M. catarrhalis, N. meningitides, and S.aureus are the most important respiratory pathogens in both severe and mild cases, causing community-acquired pneumonia, acute pharyngeal infections, sepsis, and bacterial meningitis [7,8].
Carriage or colonization of those pathogens can lead to transmission and infection. Although mere colonization does not harm the host, it could result in microbiota imbalance [9], a major risk factor for an invasion and infection of the respiratory tract and adjacent structures [10].
In order to treat febrile illness properly, the pathogens that cause febrile disease must be known. Even if the specific agent is not identified, knowing the category of the pathogen (parasitic, bacterial, or viral) is useful for deciding on treatment, particularly in resource-limited settings. Studies have shown that most (50%–75%) febrile episodes in children under 5 years of age, presented at outpatient clinics are associated with acute respiratory infections [11]. The implicated etiologic agents for most pneumonia and other upper respiratory disease are S. pneumoniae, H. influenzae, and S. aureus. The effectiveness of irrationally prescribing antibiotics to these patients should be evaluated. Lack of adequate information regarding etiological agents influences the use of inappropriate and broad-spectrum antibiotics which in turn fosters antimicrobial resistance. These further complicate the case management and its outcome. Therefore, this study aimed to determine the Oropharynx carriage rate of potentially pathogenic bacteria, associated risk factors, and antimicrobial susceptibility pattern among febrile cases in the outpatient department of Adama Hospital Medical College (AHMC).
Methods
Study design, period and setting
A cross-sectional study was conducted at AHMC, Adama city, Oromia Region, Ethiopia from November 2018 to April 2019. The city is located 99km to the east of the capital city Addis Ababa, Ethiopia. Although there are four private general hospitals in the city, AHMC is the only public teaching hospital. AHMC serves as a referral site for a catchment area population of about 5.2 million. Currently, AHMC has more than 890 workers of which 450 are health professionals and educators, with a bed capacity of 455 in eight different departments and an average outpatient flow of above 25,000.
Study population and selection
Febrile outpatients attending AHMC were included, but patients with a history of upper respiratory tract infection, recent nasal surgery, use of medications, and antimicrobial treatment (before a week of specimen collection) were excluded. In the presence of any nasal pathology/deformity in the patients, seriously sick patients were also excluded. 253 study participants were enrolled using a systematic random sampling technique in the study period.
Operational definitions
The febrile patient is a client who has a raised body temperature above the normal level (>37.5oC) measured from the axillary area.
Data collection
Information on socio-demographic and behavioral variables was collected from each study participant by face-to-face interview using a semi-structured questionnaire.
Specimen collection and processing
An oropharyngeal specimen was collected with a sterile cotton swab by rotating 4-5 times in both clockwise and counterclockwise directions before the withdrawal. After sampling, swabs were placed immediately on Amies transport medium (Oxoid, UK) and transported to the microbiology laboratory of Oromia Public Health Referral and Research Laboratory Center, Adama, Ethiopia. The swab samples were inoculated on 5% Sheep Blood Agar, Chocolate Agar, and MacConkey agar within 4 hours of collection. The anaerobic jar was used to support the growth of S. pneumonia and H. influenzae and other fastidious colonizers. All of the inoculated culture media were incubated at 37oC for 24 hrs.
Bacterial isolation and identification
Streptococcus pneumoniae isolates were identified by α-hemolytic colonies on blood agar that was exposed to optochin and bile salt. N. meningitides and H. influenzae were identified by colorless medium size colonies on Chocolate agar with X (Hemin) and V (NAD+) factors. Similarly, M. catarrhalis isolates were identified by non-hemolytic gray to white colonies on Blood agar which are catalase and oxidase test positive. S. aureus isolates were identified based on growth pattern on blood agar and fermenting mannitol on mannitol salt agar and confirmed as positive by catalase and tube coagulase test. Methicillin-Resistant S. aureus (MRSA) was identified by using Cefoxitin(30µg) as a surrogate marker for Mac A gene on S. aureus isolates as on the Clinical and Laboratory Standard Institute 2018 guideline [12].
Characterization and identification of gram-negative bacterial (Enterobacteriaceae family) isolates were done using MacConkey agar, enzymatic (catalase, oxidase, urease, and indole), and fermentative (glucose and lactose fermentation, citrate utilization, lysine decarboxylation, indole, gas, and H2S production) biochemical and motility tests. All biochemical test reagents were obtained from Oxoid, UK Company.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was done by following the Kirby Bauer disk diffusion method on Mueller Hinton agar supplemented with 5% sheep's blood (only for some that require it). Antimicrobial drugs implemented were Amoxicillin (10μg), Ampicillin (10μg), ceftazidime (30μg), cefuroxime (30μg), Nalidixic acid (30μg), Azitromycin (15μg), ceftriaxone (30μg), chloramphenicol (30μg), clindamycin (30μg), Erythromycin (15μg), tetracycline (30μg), penicillin (10 IU), ciprofloxacin (5μg), Gentamicin (10μg), Trimethoprim-sulfamethoxazole (25μg) and cefoxitin (30μg). Antimicrobial agents were selected based on clinical significance, local treatment protocol, and literature data search.
Test organism turbidity was adjusted to a 0.5 McFarland standard. Antimicrobial susceptibility testing procedure was performed and interpreted based on the Clinical and Laboratory Standard Institute 2018 guidelines [12].
Quality assurance
Data were collected by trained health professionals using a pre-tested questionnaire. To ensure data quality, all complete data sets were examined by the investigators for completeness. The completed questionnaire was cleaned, coded, and edited before data entry. Oropharyngeal swab specimens were collected, coded, labeled, and transported (Amies transport medium in Icebox) by an experienced health professional.
All reagents and culture media were prepared in accordance with standard operating procedures. Equipment operation was done according to the manufacturer’s instructions. Reference strains of S. aureus ATCC25923, S. pneumoniae ATCC49619, and H. Influenzae ATCC 49241 were used for quality control as positive controls during culture media, potency checking, gram staining, and antimicrobial susceptibility testing. A standardized bacteriological procedure was followed to maintain correct laboratory results. The sterility of the media was checked by incubating 5% of the prepared batch overnight before use.
Data processing and analysis
Data were checked, cleaned, coded, and entered into epi-info version 7 then exported to SPSS version 20 (IBM Corporation, Armonk, NY, USA) for analysis. The study population characteristics were summarized using descriptive statistics. The Oropharyngeal bacteria carrier rate and antibiotic susceptibility were presented as proportions with a 95% confidence interval. Bivariate logistic regression, independent variables with a p-value of <0.25 was considered a candidate for the multivariate logistic regression model. A multi-co-linearity test was done and the variables with variance inflation factors of greater than 10 were excluded from the multivariable logistic regression model. The association was expressed in odds ratio with 95% confidence interval and p-value <0.05 were used as cut-off points to declare significance in the final model.
Ethical considerations
An ethical approval letter was obtained from AHMC Institutional Reviewing Board. Necessary permission was obtained from the respective hospital administrative body. An information sheet was used while briefing the study participant about the objective and other related ethical issues. Written informed consent was obtained from each participant before enrolment in the study. For confidentiality, any information related to the analysis result of the study population was identified using codes, and analysis was done on data with codes. The data had been used only for the study purpose and were not disclosed to third parties.
Results
Socio-demographic characteristics
A total of 253 febrile outpatients were included in the study; of these, 151/253 (59.7%) were males. The ages of the study participants range from 1 to 75 years with a mean age of 31.1 years (SD+13.2). Most of the study participants, 117/253 (46.2%), belong to the age group <25 years of age, and 145/253 (57.3%) were from the urban area. Among the 253 study participants; 92/253 (36.4%) were smokers and 109/253 (43.1%) were high school completed students. Almost half, 121/253 (47.8%), of the study participants, had no habit to care for their oral hygiene regularly, rather they brushed their teeth sometimes and 71/253 (28.1%) never brushed their teeth (Table 1).
Oropharyngeal bacterial carriage rate
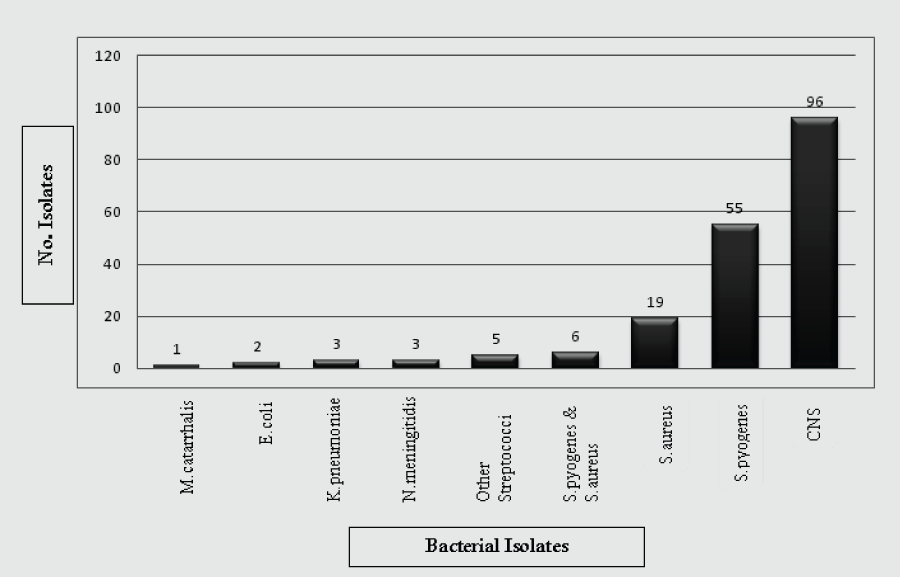
Out of 253 study participants, 190/253 (75.1%) had potential pathogenic bacteria colonization in their oropharynx. Of these, the most frequent isolate was Coagulase Negative Staphylococci (CNS) 96/190 (50.5%) followed by S. pyogenes 55(28.9%) and S. aureus 19 (10%) from which 3/19 (16%) were MRSA strains. Concurrent S. pyogenes and S. aureus colonization was found in 6/253 (2.4%). Other bacterial isolates recovered from study participants are shown in Figure 1.
Risk factors
Bivariate and multivariate logistic regressions were analyzed against socio-demographic factors. In the Bivariate analysis; independent variables (sex, age, residence, smoking status, oral hygiene, and level of education) were candidates for multivariate analysis (P <0.25). In multivariate analysis; residence [AOR= 2.06; 95% CI= (1.31-8.97), P= 0.026], smoking [AOR= 4.24; 95% CI= (3.65-15.23), P= 0.029], and oral hygiene [AOR= 3.29; 95% CI= (3.10-9.437), P= 0.012] were significantly associated with oropharyngeal bacterial colonization (Table 2).
Antimicrobial susceptibility patterns
The antimicrobial susceptibility test result showed that the majority of CNS were resistant to penicillin 77 (80.2%), erythromycin 63 (65.4%), and cotrimoxazole 60 (62.5%). Likewise, S. aureus isolates were also resistant to β-lactam antibiotics and cotrimoxazole. However, most of the S. aureus isolates were susceptible to chloramphenicol 18(94.7%), clindamycin 17 (89.5%), and ciprofloxacin 16 (84.2%). About, 16% (3/19) of S. aureus isolates were MRSA strains. The majority of S. pyogenes isolates were resistant to Ampicillin 35 (63.6%) and tetracycline 40 (72.7%). From gram-negative isolates, E. coli and K. pneumoniae showed 100% resistance to amoxicillin, ampicillin, and cotrimoxazole. However, the majority of these gram-negative isolates were susceptible to ceftazidime, ceftriaxone, cefuroxime, and azitromycin (Table 3).Regarding multiple drug-resistant isolates, 9 K. pneumoniae, 8 S. aureus, and 6 CNS isolates showed resistance for nine different antibiotic classes (Table 4).
Discussion
Oropharyngeal asymptomatic carriage is a predisposing factor for developing invasive and non-invasive diseases, and it could serve as a source for hospital-associated infection and even to the community. The oropharyngeal bacteria carriage rate (190/253) obtained from the current study is higher than the study conducted in Brazil (55.9%) [13] and BahirDar, Ethiopia (51%) [12]. The most frequent oropharyngeal bacteria isolate was CNS which accounted for 37.9%) of the total isolates. Again, this finding was higher than studies conducted in India (14.3%) [14] and (15.4%) [15]. The difference in the carriage rate may be due to the variation of the study participants as our participants were febrile patients.
The second predominant isolate was S.pyogenes which accounted for 21.7% of all isolates. It is comparable to a study done in Uganda (16%) [16]. However, it is higher than studies done in Chicago (9.6%) [17], Netherlands (6.3%) [18], Nepal (5.3%) [10] and Jimma, Ethiopia (11.3%) [19].
The Oropharyngeal colonization rate of S. aureus was 7.5%. The finding is comparable (10.3%) with a study done in Gondar, Ethiopia [20], but lower than studies reported in Chicago (26.2%) [17], Italy (25.9%) [21], Nepal (14.7%) [10] and Senegal (56.1%) [22] and BahirDar, Ethiopia(29%) [12]. The variation of carriage rate of oropharyngeal bacteria among these studies might be due to differences in the characteristics of the study population, quality of sampling, geographical distribution, and seasonal effect.
The finding from the current study showed that being urban had more chances to have oropharyngeal bacterial carriage than being rural. The possible reason might be sweet foods item linked to urbanization, which increases oral bacterial growth. Pharyngeal bacterial colonization was also significantly higher among smokers than non-smokers. The possible reason might be impairment immunity related to smoking. The current study also revealed that the proportion of oropharyngeal bacterial carriage was significantly higher in participants who had no regular trend of brushing their teeth compared to others. The finding is in agreement with other studies done in Indonesia [23] and Ethiopia [12].
Regarding antimicrobial susceptibility pattern result, penicillin was the least effective antibiotic with 80.2% and 84.2% revealed resistance for CNS and S.aureus respectively. This is in line with various studies which were conducted in China (87.5%) [24], Ghana (95.1%)[25], Gondar, Ethiopia (99.3%)[26], Bahir Dar, and Ethiopia(84.4%) [12]. On the other hand, S. aureus isolates were susceptible to chloramphenicol 18(94.7%), clindamycin 17(89.5%), ciprofloxacin 16(84.2%), and cefoxitin 16(84.2%). This finding is in agreement with studies conducted in Ethiopia [26,27].
Streptococcus pyogenes revealed resistance to tetracycline (72.7%), ampicillin (63.6%), and erythromycin (43.6%) in the present study. This finding is higher than the study conducted in Germany (12.7%) [28] and (9.8%) [29] for erythromycin, and Gondar, Ethiopia (59.4%) [30] for tetracycline. However, it is lower than the study conducted in Senegal which showed (100%) resistance for tetracycline [31].
In this study amoxicillin, ampicillin, and cotrimoxazole showed 100% resistance for E. coli and K. pneumoniae. On the other hand, the majority of antibiotics including ceftazidime, ceftriaxone, cefuroxime, and Azitromycin were found to be effective. This finding is supported by studies done in India [32] and Bahir Dar, Ethiopia [33].
Furthermore, the majority of Oropharyngeal bacteria isolates were resistant to three and more treatment options antibiotics. Nine K. pneumoniae, 8 S. aureus, and 6 CNS isolates showed resistance for nine antibiotic classes each (Table 4). The differences in susceptibility pattern of the isolates might be associated with frequent and irrational use of antibiotics, easy availability of drugs, self medications, illegal purchase, and being broad-spectrum nature of the drugs may lead to misuse of antibiotics.
Conclusion
The current study revealed that the overall rate of oropharyngeal bacterial colonization is high (75.1%) in AHMC. Living in urban, tobacco smoking, and poor oral hygiene are risk factors associated with the high oropharyngeal pathogenic bacterial carriage. Antimicrobial susceptibility pattern showed that different classes of drugs like penicillin, tetracycline, ampicillin, erythromycin are resistant especially for gram-positive bacteria. Therefore, identifying oropharyngeal bacteria and risk factors as well as determining the antimicrobial susceptibility pattern of the isolates are important components for the management and prevention of these infectious diseases. Besides, it is recommended that clinicians give due emphasis to oropharyngeal bacteria in febrile patients and health facilities provide health education to the customers.
Declarations
Ethics approval and consent to participate: An ethical approval letter was obtained from AHMC Institutional Reviewing Board. Necessary permission was obtained from the respective hospital administrative body. An information sheet was used while briefing the study participant about the objective and other related ethical issues. Written informed consent was obtained from each participant before enrolment in the study. For confidentiality, any information related to the analysis result of the study population was identified using codes, and analysis was done on data with codes. The data had been used only for the study purpose and did not disclose to third parties.
Availability of data and material: The datasets generated and/or analyzed during the current study are not publicly available. Sharing of data was not included in the approval from the ethics committee but is available from the corresponding author on a reasonable request.
Funding: Adama Hospital Medical College funds this research work. The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation.
Authors' contributions
MTW conceived the study and was involved in the study design, reviewed the article, analyzed, reported writing, and drafted it. TSS, TEC, HLA, BY, MT, KJ, and WB were contributed to data analysis, report writing, drafted the manuscript, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
The authors acknowledge Adama Hospital Medical College and Armauer Hansen Research Institute for funding this study; and study participants for their cooperation.
- Humphreys GJ, McBain AJ (2013) Continuous culture of sessile human oropharyngeal microbiotas. J Med Microbiol 62: 906-916. Link: https://bit.ly/3hHHTxd
- Adegbola RA, DeAntonio R, Hill PC, Roca A, Usuf E, et al. (2014) Carriage of Streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PloS One 9: e103293. Link: https://bit.ly/3Mu1ezT
- Bissong M, Fon P, Kamga F, Akenji T (2014) Microbiological Profile of Oral Infections in Diabetic Patients and Non-Diabetic Controls in SouthWest, Cameroon. Afr J Clin Experiment Microbiol 15: 138-143. Link: https://bit.ly/3sNbziB
- Dunne EM, Murad C, Sudigdoadi S, Fadlyana E, Tarigan R, et al. (2018) Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: a cross-sectional study. PloS One 13: e0195098. Link: https://bit.ly/3HNTFka
- Bhuyan GS, Hossain MA, Sarker SK, Rahat A, Islam MT, et al. (2017) Bacterial and viral pathogen spectra of acute respiratory infections in under-5 children in hospital settings in Dhaka city. PloS One 12: e0174488. Link: https://bit.ly/3Ciex1G
- Lee CJ, Sankaran S, Mukherjee DV, Apa ZL, Hafer CA, et al. (2011) Staphylococcus aureus oropharyngeal carriage in a prison population. Clin Infect Dis 52: 775-778. Link: https://bit.ly/3MthNvZ
- Jounio U (2012) Oropharyngeal Carriage of Respiratory Bacteria Among Military Conscripts: University of Oulu.Link: https://bit.ly/3KmQRMB
- Sheikhi R, Amin M, Rostami S, Shoja S, Ebrahimi N (2015) Oropharyngeal colonization with Neisseria lactamica, other nonpathogenic Neisseria species and Moraxella catarrhalis among young healthy children in Ahvaz, Iran. Jundishapur J Microbiol 8: e14813. Link: https://bit.ly/3MtE4tp
- Chochua S, D'Acremont V, Hanke C, Alfa D, Shak J, et al. (2016) Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PloS One 11: e0167725. Link: https://bit.ly/3sNUKnW
- Thapa S, Gokhale S, Sharma AL, Sapkota LB, Ansari S, et al. (2013) Burden of bacterial upper respiratory tract pathogens in school children of Nepal. BMJ Open Respir Res 4: e000203. Link: https://bit.ly/3KhpW4G
- Petti S, Sofan AA, Messano GA (2013) Streptococcus pneumoniae carriage rate in healthy preadolescent dental patients. Acta Stomatologica Naissi 29: 1249-1254. Link: https://bit.ly/36XWuSU
- Mulu W, Yizengaw E, Alemu M, Mekonnen D, Hailu D, et al. (2018) Pharyngeal colonization and drug resistance profiles of Morraxella catarrrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae among HIV infected children attending ART Clinic of Felegehiwot referral hospital, Ethiopia. PloS One 13: e0196722. Link: https://bit.ly/3CiJN0L
- Ternes YM, Lamaro-Cardoso J, André MCP, Pessoa VP, da Silva Vieira MA, et al. (2013) Molecular epidemiology of coagulase-negative Staphylococcus carriage in neonates admitted to an intensive care unit in Brazil. BMC Infect Dis 13: 572. Link: https://bit.ly/34iNIxR
- MIR AM, Srikanth (2013) Prevalence and Antimicrobial Susceptibility of methicillin resistant staphylococcus aureus and coagulase-negative staphylococci in a tertiary care hospital. Asian J Pharm Clin Res 6: 231-234. Link: https://bit.ly/3hLPnPK
- Rahman M, Islam MN, Islam MN, Hossain MS (2015) Isolation and identification of oral bacteria and characterization for bacteriocin production and antimicrobial sensitivity. Dhaka Univ J Pharm Sci 14: 103-109Link: https://bit.ly/3Cswn2k
- Nayiga I, Okello E, Lwabi P, Ndeezi G (2017) Prevalence of group a streptococcus pharyngeal carriage and clinical manifestations in school children aged 5–15 yrs in Wakiso District, Uganda. BMC Infect Dis 17: 248. Link: https://bit.ly/3HM5es3
- Levy R, Leyden J, Margolis D (2005) Colonisation rates of Streptococcus pyogenes and Staphylococcus aureus in the oropharynx of a young adult population. Clin Microbiol Infect 11: 153-155. Link: https://bit.ly/35Yt9aF
- Liassine N, Gervaix A, Hegi R, Strautmann G, Suter S, et al. (1999) Antimicrobial susceptibility of bacterial pathogens in the oropharynx of healthy children. Eur J Clin Microbiol Infect Dis 18: 217-220. Link: https://bit.ly/3Cig6wC
- Tesfaw G, Kibru G, Mekonnen D, Abdissa A (2015) Prevalence of group A β-haemolytic Streptococcus among children with pharyngitis in Jimma town, Southwest Ethiopia. Egyptian Journal of Ear, Nose, Throat and Allied Sciences 16: 35-40. Link: https://bit.ly/3CkTRGu
- Assefa A, Gelaw B, Shiferaw Y, Tigabu Z (2013) Nasopharyngeal carriage rate and antimicrobial susceptibility pattern of potential pathogenic bacteria among pediatric outpatients at Gondar University teaching hospital, Ethiopia. J Infect Dis Ther 21: 109. Link: https://bit.ly/3IRKgcB
- Esposito S, Terranova L, Zampiero A, Ierardi V, Rios WP, et al. (2014) Oropharyngeal and nasal Staphylococcus aureus carriage by healthy children. BMC Infect Dis 14: 723. Link: https://bit.ly/3HNTghE
- Fall C, Richard V, Dufougeray A, Biron A, Seck A, et al. (2014) Staphylococcus aureus nasal and pharyngeal carriage in Senegal. Clin Microbiol Infect 20: O239-O241. Link: https://bit.ly/3IR2L0W
- Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, et al. (2014) Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PloS One 9: e91999.Link: https://bit.ly/3i1Dc1v
- Zhu H, Wang A, Tong J, Yuan L, Gao W, et al. (2015) Nasopharyngeal carriage and antimicrobial susceptibility of Haemophilus influenzae among children younger than 5 years of age in Beijing, China. BMC Microbiol 15: 6. Link: https://bit.ly/3CrG0hB
- Eibach D, Nagel M, Hogan B, Azuure C, Krumkamp R, et al. (2017) Nasal carriage of Staphylococcus aureus among children in the Ashanti Region of Ghana. PloS One 12: e0170320. Link: https://bit.ly/3vKEy8A
- Tigabu A, Tiruneh M, Mekonnen F (2018) Nasal carriage rate, antimicrobial susceptibility pattern, and associated factors of Staphylococcus aureus with special emphasis on MRSA among urban and rural elementary school children in Gondar, Northwest Ethiopia: A comparative cross-sectional study. Adv Prev Med 2018: 9364757. Link: https://bit.ly/3tzn6kW
- Reta A, Gedefaw L, Sewunet T, Beyene G (2015) Nasal carriage, risk factors and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus among school children in Ethiopia. J Med Microb Diagn 4: 1. Link: https://bit.ly/35yWDfj
- Arvand M, Hoeck M, Hahn H, Wagner J (2000) Antimicrobial resistance in Streptococcus pyogenes isolates in Berlin. J Antimicrob Chemother 46: 621-624. Link: https://bit.ly/35BB9hU
- Imöhl M, van der Linden M (2015) Antimicrobial susceptibility of invasive Streptococcus pyogenes isolates in Germany during 2003-2013. PloS One 10: e0137313. Link: https://bit.ly/3vOJCJc
- Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F (2017) Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol 2017: 8953829. Link: https://bit.ly/3Mu7t6Y
- Camara M, Dieng A, Boye CSB (2013) Antibiotic susceptibility of streptococcus pyogenes isolated from respiratory tract infections in dakar, senegal. Microbiol Insights 6: 71-75. Link: https://bit.ly/34mOHxc
- Manikandan C, Amsath A (2013) Prevalence and distribution of bacteria and fungi isolated from patients with urinary tract infection in pattukottai, tamilnadu, India. Int J Pure Appl Zool 1: 213-222. Link: https://bit.ly/3KlbtEN
- Kibret M, Abera B (2011) Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr Health Sci 11: 40-45. Link: https://bit.ly/3sPbsmY
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
